Kanteron team introduces Panelmaps at European Society of Human Genetics meeting in Sweden
Kanteron Systems presents its latest R&D development in NGS and CNV analysis application, Panelmaps, at the European Society of Human Genetics (ESHG19) annual meeting in Gothenburg (Sweden).

During the past 6 month Kanteron Systems has been working closely with the University of Cambridge’s clinical genomics initiative OpenCB, contributing code and clinical know-how. This software is currently being used by top pharmaceutical companies and large genomics projects like Genome England, where optimized systems that can handle millions of genomes efficiently are needed.
Led by our Director of Bioinformatics, Dr. Pablo Marin along other members of his team and his doctoral candidate mathematician Josemi Juanes from Dr. Javier Chaves’ laboratory at the Healthcare Research Institute INCLIVA1, the new software has been developed in collaboration with OpenCB, MGviz and Seqplexing. E-poster E-P16.03 “PanelMaps: a genome-scale coverage QC and CNV advisor web application”.
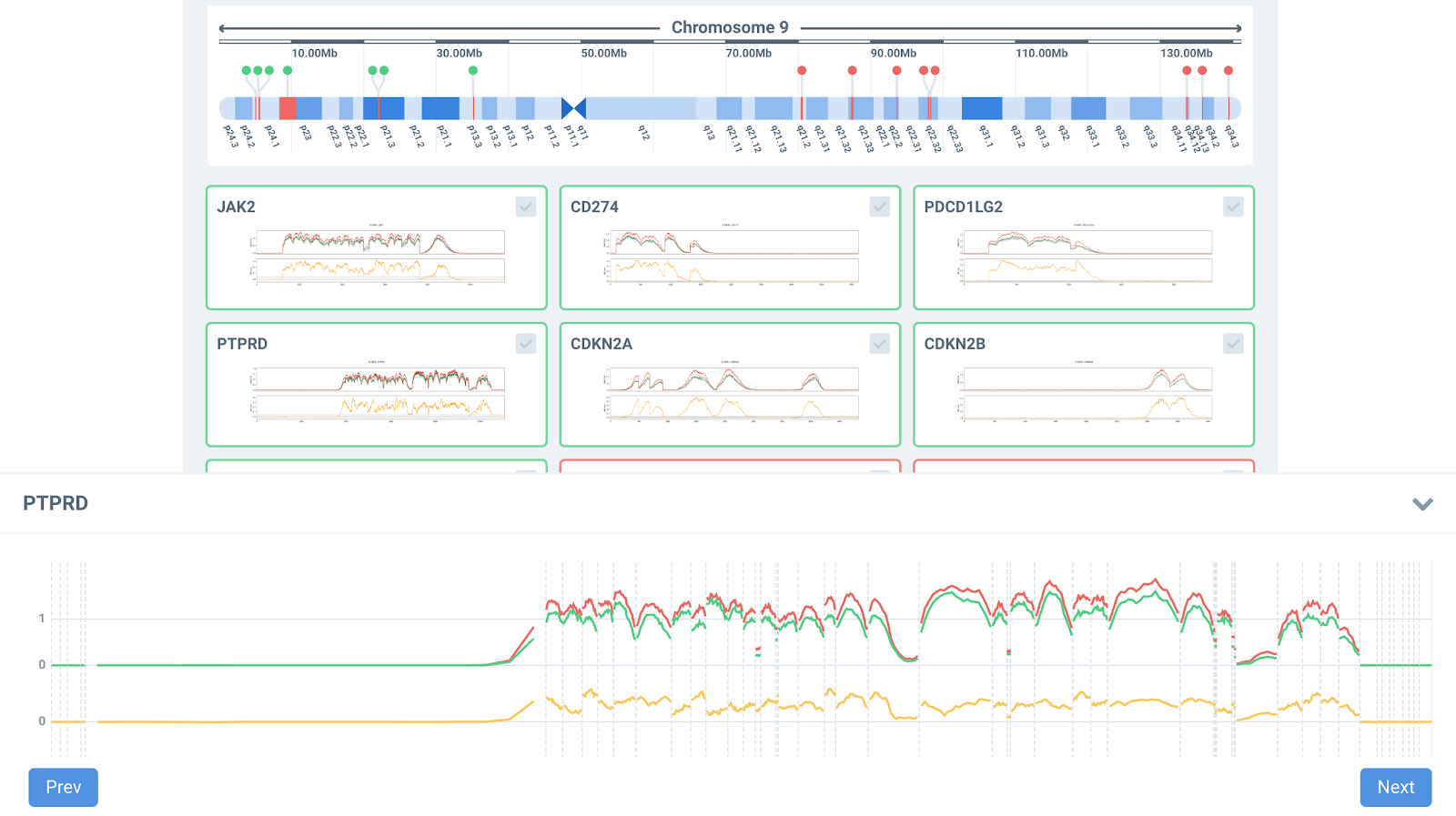
Panelmaps uses adaptative machine learning algorithms for normalizing cancer data, avoiding putative regions with CNV for normalizing the data. It also computes LOH and provides a CNV advisory score, along with the values of the CNV programs run by the user.
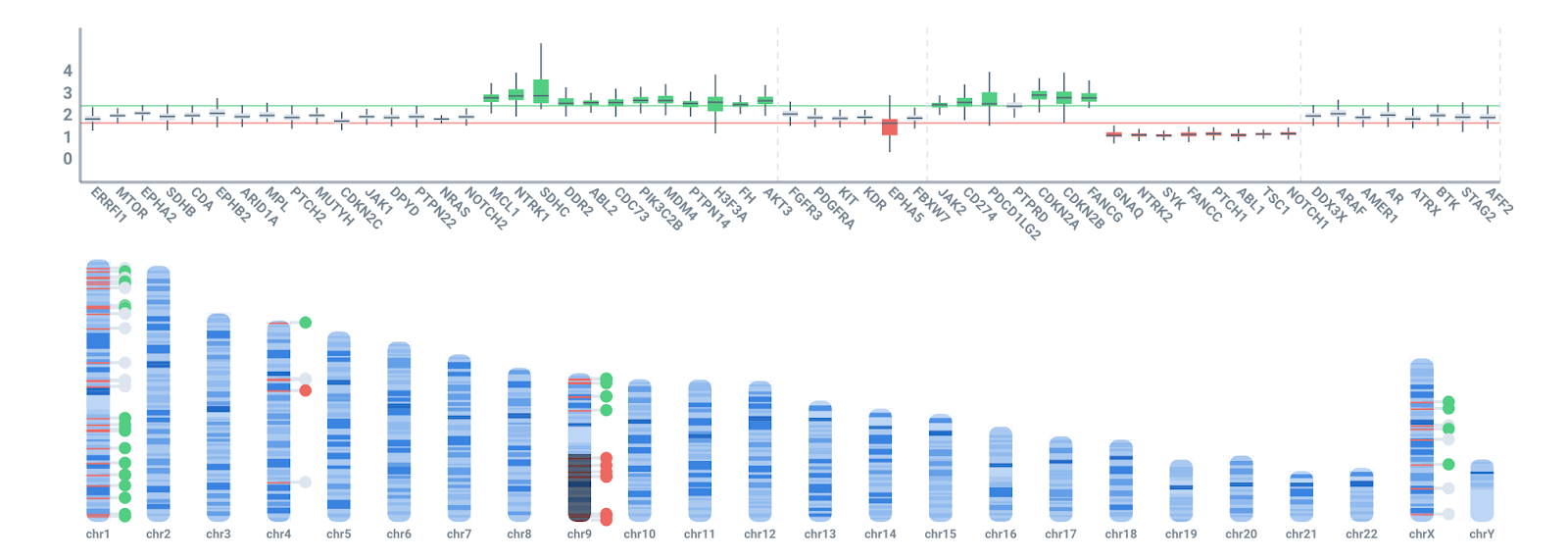
Panelmaps allows to quickly visualize the results of your NGS experiments and helps explore CNVs genomewide. The software has an advisor mode that shows the copy number in gene summaries along a karyoview, showing also the LOH (dark areas in the ideogram) in the CNV region with probes.
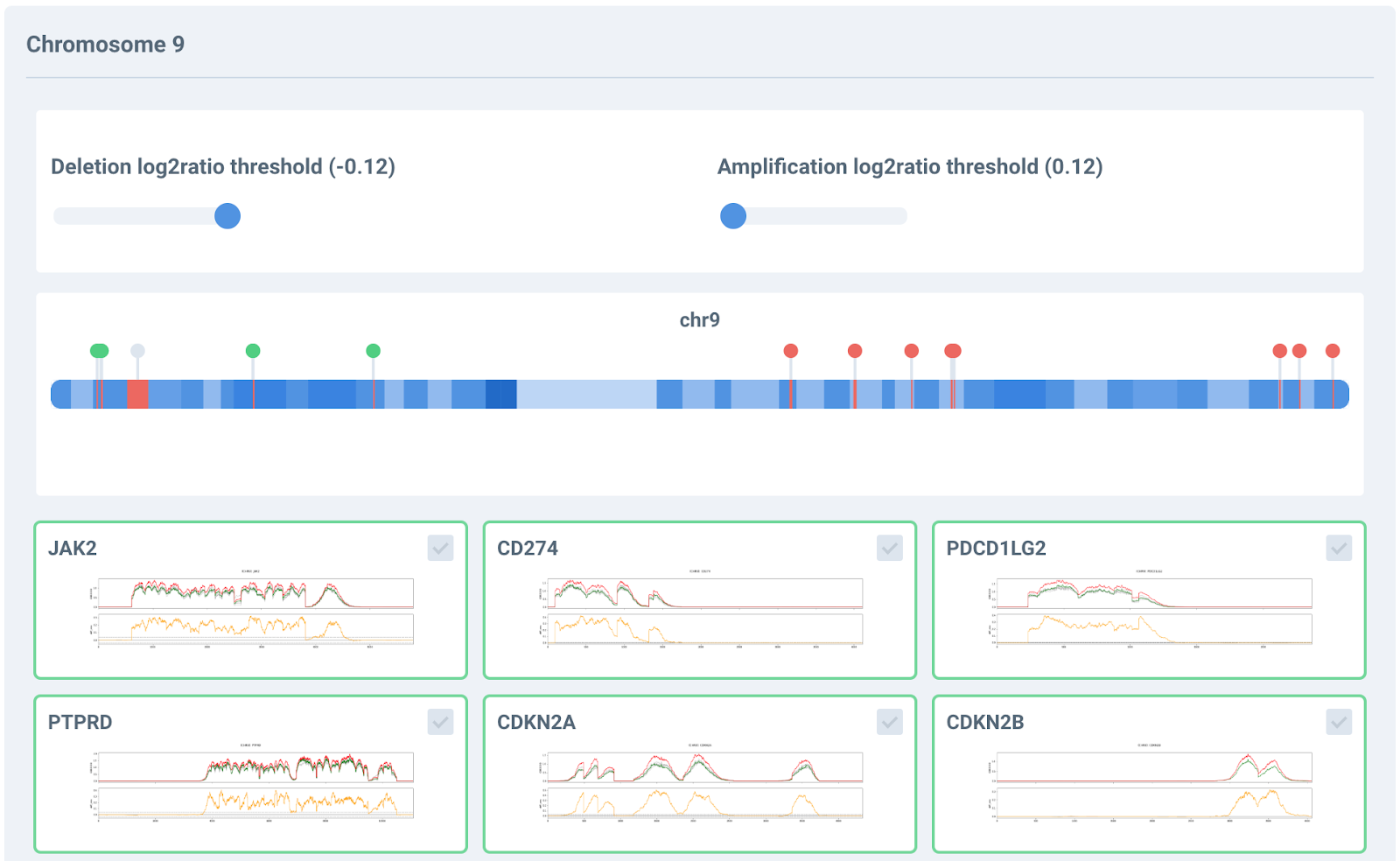
Panelmaps facilitates interactive exploration of all CNV signals, allowing the selection of different thresholds for duplication and deletion events. It also computes germline contamination and helps annotate CNVs in the clinical variant storage.
At Kanteron Systems we’re proud of the R&D work our Bioinformatics department is producing, in collaboration with other institutions: open-science and open-source. This research is later converted, with the help of our Software Development department, into Precision Medicine solutions available to clinicians worldwide.
Great job, team!
Scholarship by Instituto de Salud Carlos III through the project “IFI15/00138” (Co-funded by European Regional Development Fund). ↩︎
